library(tidyverse)
library(prospectr)
load("C:/BLOG/Workspaces/NIR Soil Tutorial/post5.RData")As in previous posts, we are going to follow the Soil spectroscopy training material and we are going to use the reference material provided.
You can find several posts about MSC in: nir-quimiometria.blogspot.com.
You can also read the explanation of the reference material in the Soil spectroscopy training material.
As always we load our data and the packages we need:
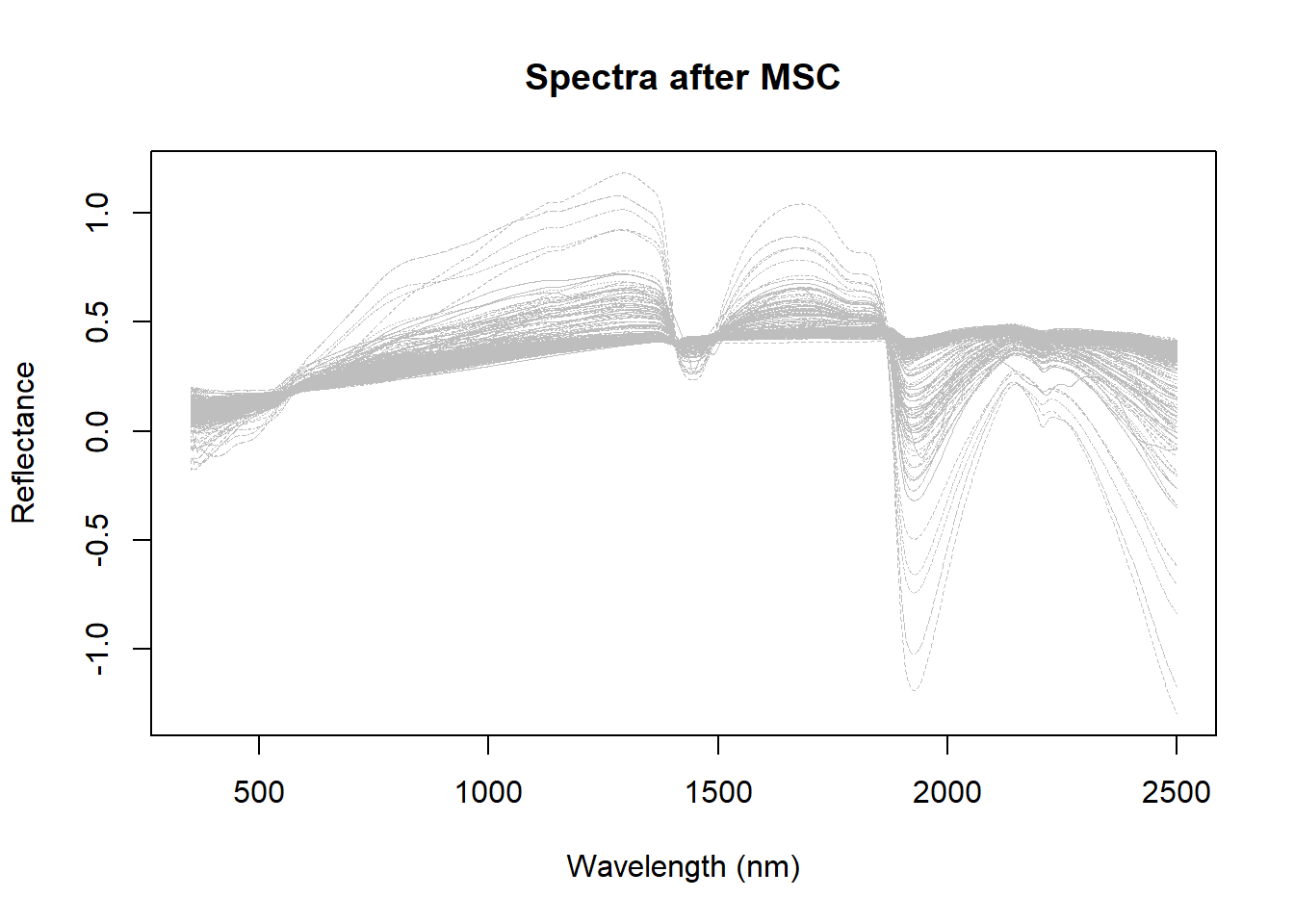
msc_spectra <- msc(dat$spc, ref_spectrum = colMeans(dat$spc))Let´s use now classical R to see the spectra:
matplot(colnames(dat$spc),
t(msc_spectra),
type = "l",
col = "grey",
lwd = 0.5,
xlab = "Wavelength (nm)",
ylab = "Reflectance",
main = "Spectra after MSC"
)
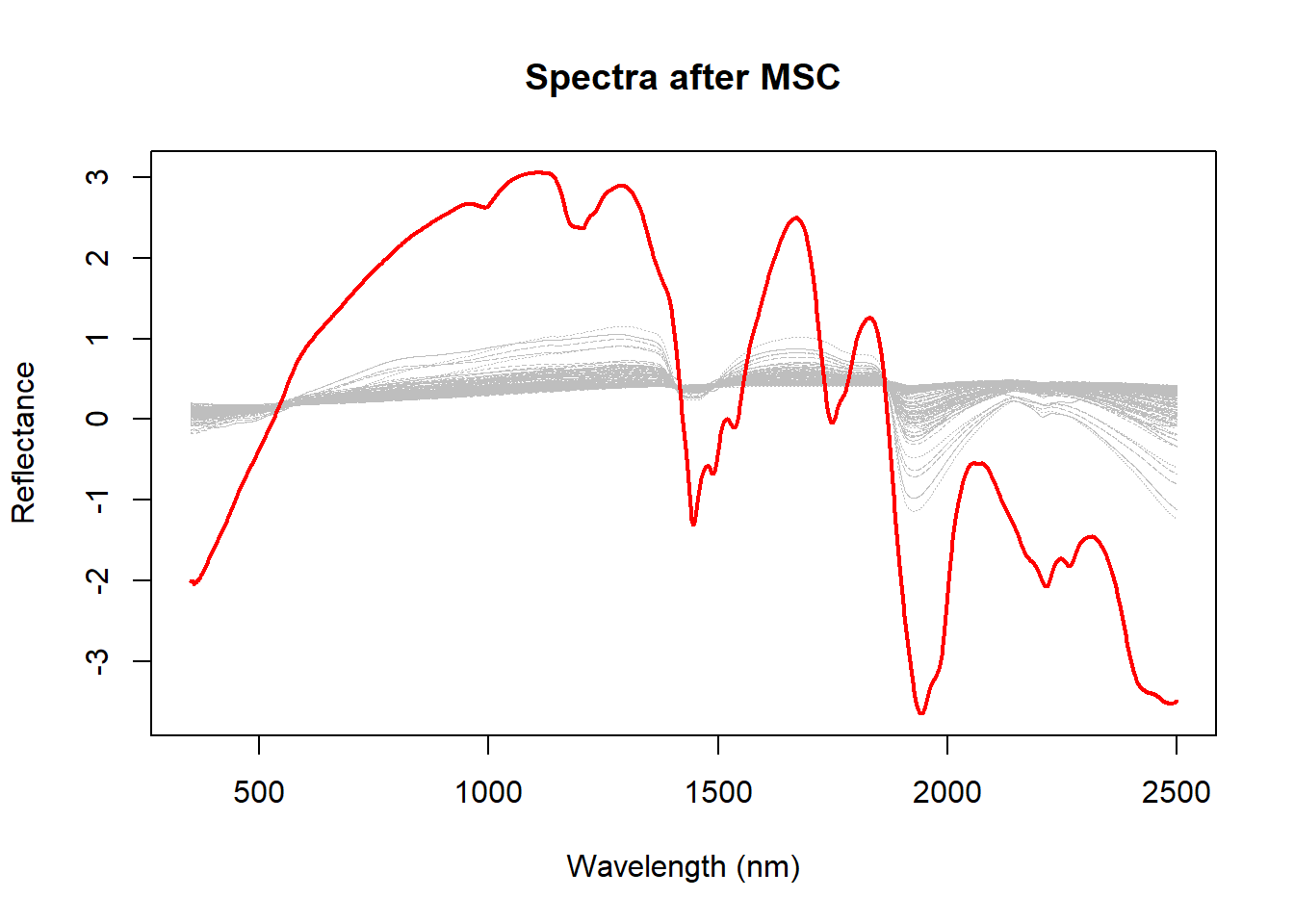
As we can see the scatter effect is minimized, but at the same time we see a sample that is very different from the others. One idea can be to remove this sample and recalculate the MSC.
# Compute the mean reflectance per sample
msc_mean_reflectance <- rowMeans(msc_spectra)
# Get the sample index with the highest mean reflectance
msc_out_sample_index <- which.min(msc_mean_reflectance)
msc_out_sample_index[1] 102matplot(colnames(dat$spc),
t(msc_spectra),
type = "l",
col = "grey",
lwd = 0.5,
xlab = "Wavelength (nm)",
ylab = "Reflectance",
main = "Spectra after MSC"
)
# Highlight the selected spectrum in red
lines(
x = colnames(dat$spc),
y = msc_spectra[102, ],
col = "red",
lwd = 2
)
Remove this sample and calculate the MSC again:
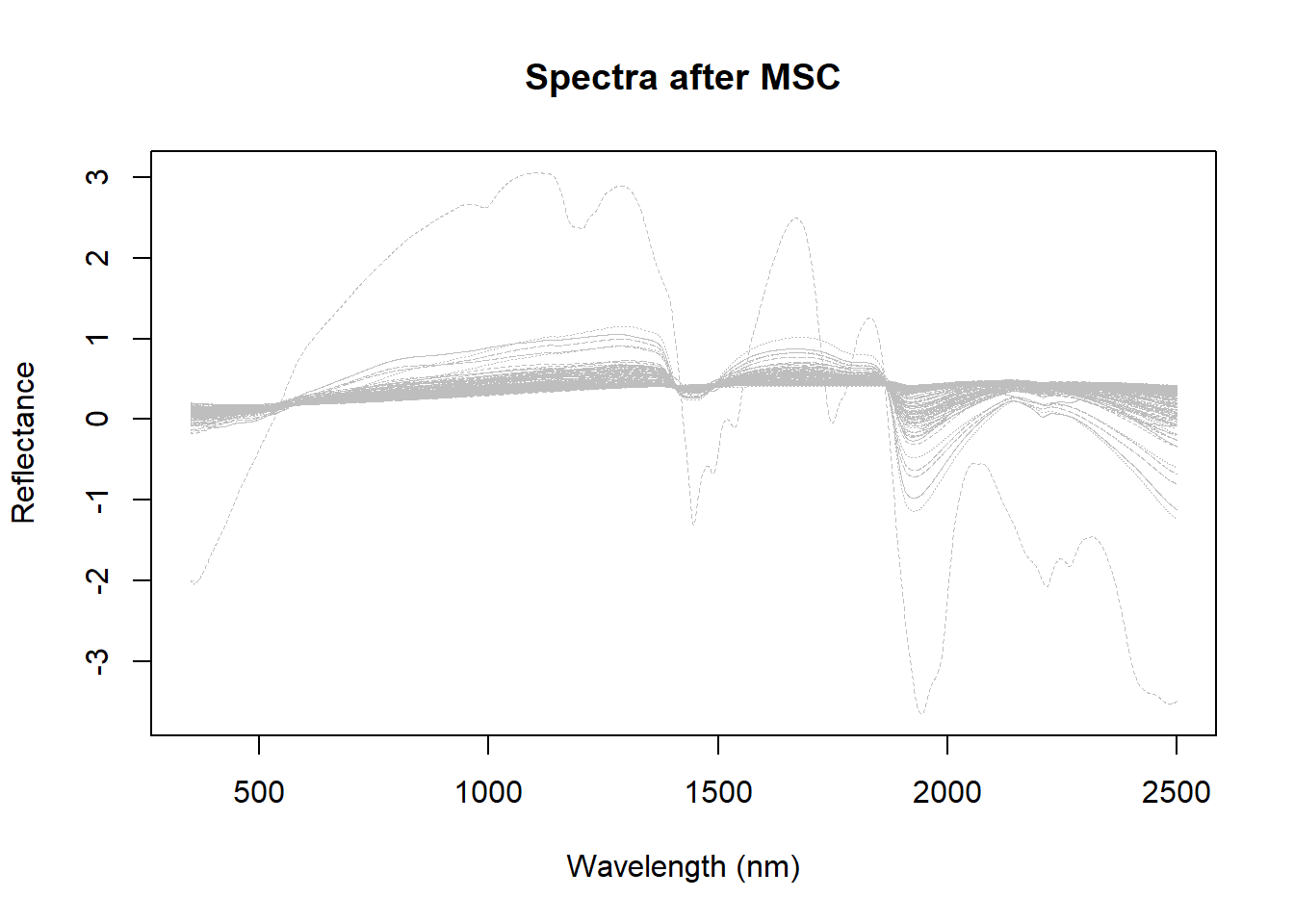
msc_spectra <- msc(dat$spc[-msc_out_sample_index, ], ref_spectrum = colMeans(dat$spc[-msc_out_sample_index, ]))
matplot(colnames(dat$spc),
t(msc_spectra),
type = "l",
col = "grey",
lwd = 0.5,
xlab = "Wavelength (nm)",
ylab = "Reflectance",
main = "Spectra after MSC"
)